Sparks of Diffusion
I've been focusing my attention on the ISLES24 dataset for close to two years now, using it as a test base for various experiments that have gotten me accustomed to the multi-modal CT Scan images and derivatives.
Another blog post is destined to contain a proper writeup of my experiences with this thusfar, but today, I want to celebrate a small win that has been a long time coming.
In the course of building my personal medical image segmentation toolkit capabilities to study ISLES24, I've been working on expanding into Diffusion models. Specifically, the MedSegDiff model.
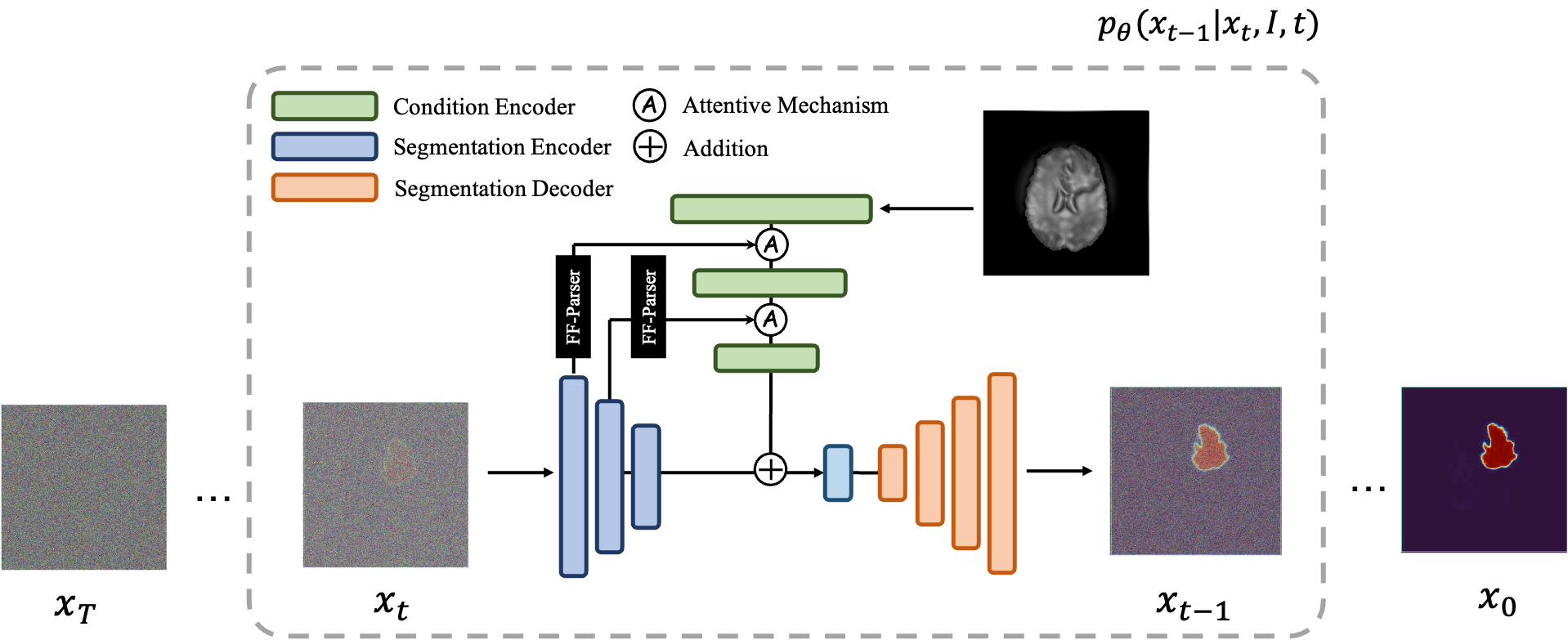
I had initially started in hopes of making the v2 model work on my problem, but after quite some messing around with the codebase and setting it up to work with ISLES24 loading, only then did I discover that the v2 model would not appear to be implemented in the official github repository of: https://github.com/ImprintLab/MedSegDiff/
The implementation itself was for me as an absolute beginner in the field of diffusing anything, quite overwhelming. It is now that after a bit around the block that I have come to understand the repo as standing on the shoulders of the giant
https://github.com/openai/improved-diffusion/tree/main/improved_diffusion
I will be getting to this repository shortly.
I found various issues with the repo that I elected to begin with a base implementation from https://github.com/deepmancer/medseg-diffusion
This was an juypter notebook that I broke down into components classes and files, and started adding experimental structures that would allow me to conduct configurable, extensible research into using this model for the ISLES24 Dataset as well as ported over my ISLES24 manipulation code from my existing SwinUNetR experiments.
My first experiments were beleaguered by various issues, as I got to discover the technique, I came to understood that the medseg-diffusion code relied on Denoising Diffusion Probabilistic Models (DDPM) reverse diffusion. This proved to be a learning experience when, in a bid to enhance segmentation mask clarity, I increased the number of reverse diffusion timesteps from 100 to 500, discovering that increasing reverse-diffusion timesteps does in fact, not come without a cost 😄
This is where I came to learn of Denoising Diffusion Implicit Models (DDIM) diffusion, and how through deterministic diffusion, steps could be jumped, as opposed to having to calculate the reverse-diffusion process for each timestep!
It is here that I learned of OpenAI's github Diffusion repositories:
https://github.com/openai/guided-diffusion/ and https://github.com/openai/improved-diffusion. In hindsight, now that I'm writing up this post, I've come to immediately recognize how the official MedSegDiff repo builds in the code structure left by the guided diffusion repo, adding dataset loaders, a new model structure and relevant changes to get these to click, but otherwise, this was the same repo.
Lucky for me, I came to recognize the utility of these codebases and in hopes of upgrading my codebase with DDIM capabilities, I too grafted the code improved-diffusion codebase into mine as a package. It felt like Megamant NT Warrior when you'd slot in an upgrade chip and suddenly your warrior has all these extra fancy capabilities!
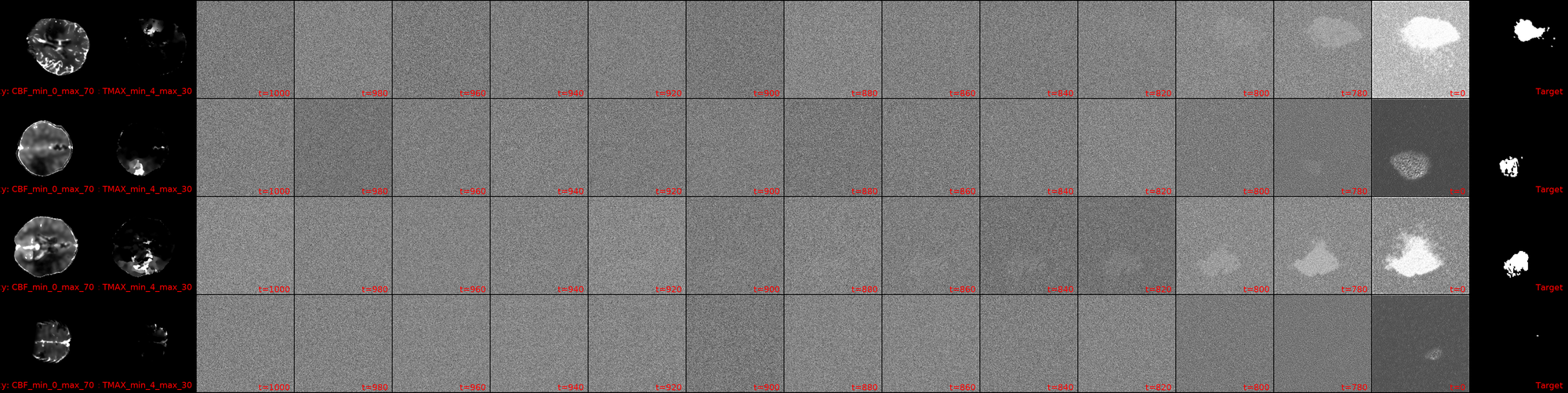
It is at this point that the post's title image comes into play. This is my first training run of a large model (specifics will be discussed in a later, probably next, post) with DDIM reverse diffusion, with 1000 timesteps, but a respacing of the timesteps to 250 (reducing the compute load by 4x). The image, for now, contains a bug from the days of having trained using only 100 timesteps, but this will be fixed shortly (in case you happen to read this in the interim, 👋). But what you can see, is how I have image masks generated via diffusion that dont look absolutely useless! Now, there's still a lot to tune here: the images being fed, augmentations, the model architecture can and will be updated, the learning rate annealing parameters tuned and the diffusion parameters as well, but for the first time, we have Sparks of Diffusion. ⚡
Watch this space.
--------------------
23.11. Update:
I couldnt bring myself to delete the older, blog post title picture, so instead, Im adding the updates additively, rather than overwriting. I got a slight bit emotional seeing the progress...
I dont know whether you can see it, but I can. This is a snapshot from one of my current crop of diffusion models! First out the gate, lets ground our expectations: the Dice scores are still s*it, with the maximum -current- score being 6.5%.
That being said, I'm still going to shamelessly showcase a few cherry picked samples that show how we actually now have clean blacks at the end of our diffusion inference! We also have segmentation masks that (to some degree) match the actual lesions in shape/location.
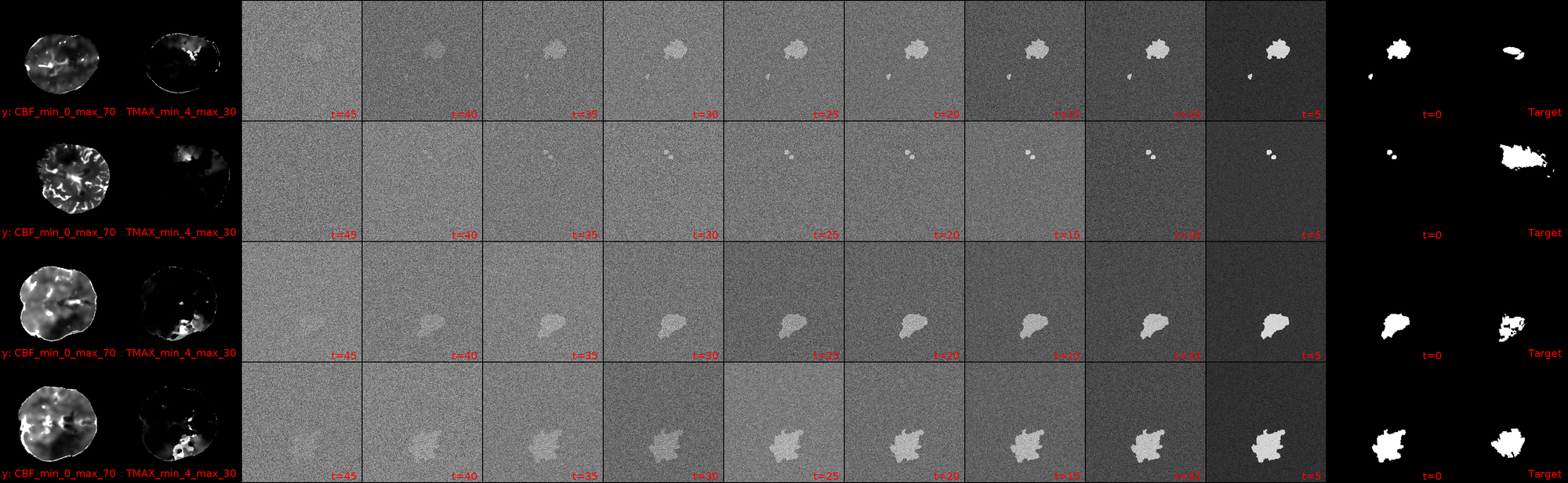
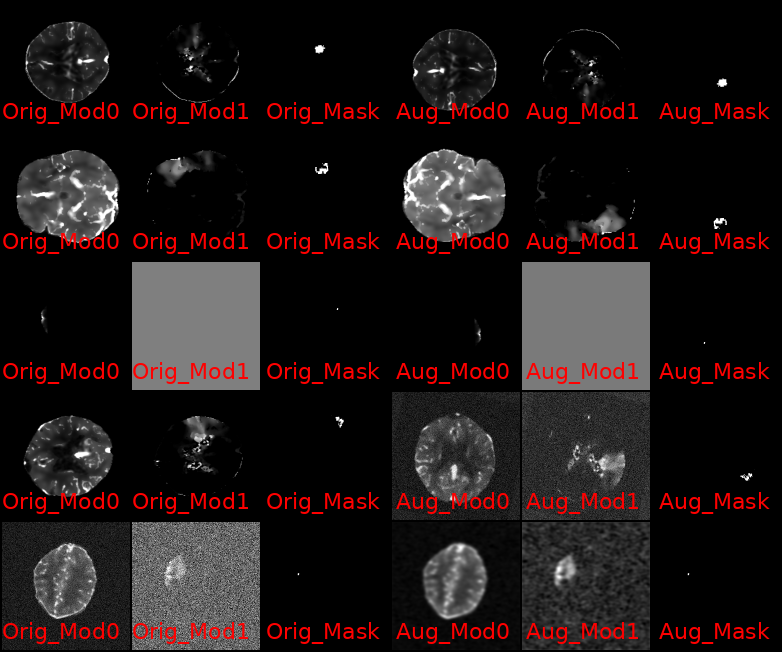
Among the advancements that I've added to the codebase since, are the following few that I want to highlight:
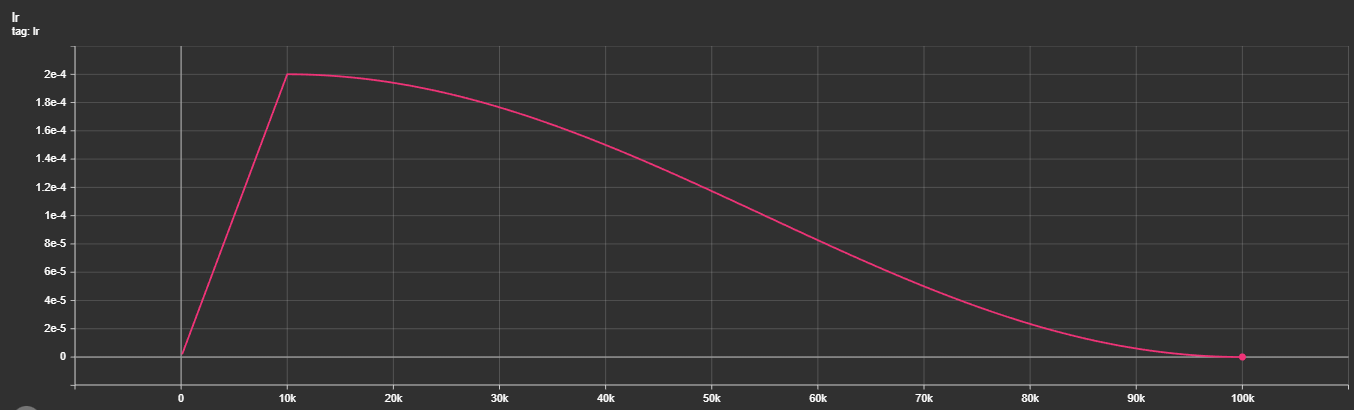
The integration of Multi-Task Losses hasn't been exactly the smoothest, take a look:
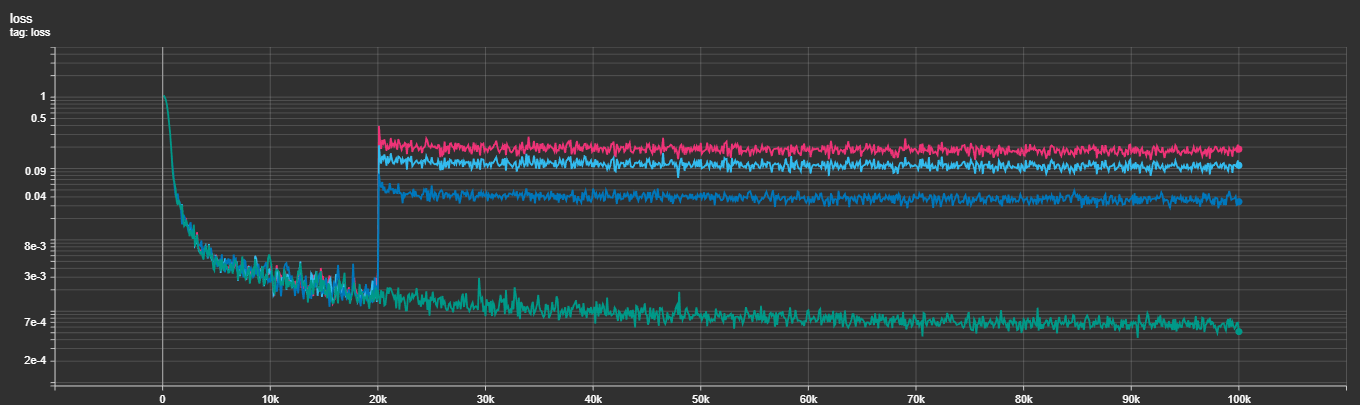
As you can see in the figure above, it's very obvious that adding the other losses (at several different weightings) results in the loss jumping multiple orders of magnitude and never quite declining the same. I'm still experimenting with this, with ideas being learned weightings to the losses, different warm-in strategies and others. It's still open.
I've started added gradient accumulation, I'm currently using a physical (on-GPU) batch size of 4. This apparently could be a reason for my metrics not trending upwards beyond what appear to be a random oscillation around the 5.0-6.5% Dice.
It stands that I could experiment around a bit with gradient accumulation till I figure out if indeed that's the source of my issues. If it is, I'll start scaling my runs to actually run the necessary batch size in a single go on multiple GPUs, but I need to see some indication.
The direction forward is still open: I'm experimenting with the training hyperparameters (learning rate scheduler, rate, batch size, augmentation) as well as with the model architectural hyperparameters and loss functions while currently remaining bound to the MedSegDiff architecture that we started our experiments out with. I'm planning to expand to other architectures the moment I start catching a signal and figuring out what combination of parameters would lead to results starting to catch on.
As always, watch this space 😃